Detection of H3K27M mutation in cases of brain stem subependymoma
2019-07-08 16:13 作者:三博腦科醫(yī)院
Kun Yao, Zejun Duan, Yin Wang, Mingshan Zhang, Tao Fan, Bin Wu, Xueling Qi
Abstract
Subependymomas are rare, slow-growing, grade I glial tumors of the central nervous system (CNS). Recently, diffuse midline gliomas with muta- tions in the H3.1 or H3.3 genes at the position of amino acid 27, resulting in the replacement of lysine by methionine (K27M), which were defined as the new grade IV entity. As H3K27M mutations have been reported in midline gliomas, gangliogliomas and pilocytic astrocytomas, whether they occur in midline subependymomas has been unclear. We were to determine whether any such mutations can be found in them and to analyze the prognostic relevance of any such mutations in subependymo-mas. Four subependymomas, all in the brainstem, harbored H3K27M mutations; No such mutation was found in any of the subependymomas from other locations. The mutations were identified by immu-nohistochemical stains and confirmed with Sanger sequencing. The median follow-up of the patients with the mutations in their tumors was 3.2 years, and 3 are still alive, having received no adjuvant therapy.We demonstrate that H3K27M mutation can occur in brainstem subependymomas, despite the presence of H3K27M mutation, these cases should not be diag- nosed or treated as grade IV tumors. Because they showed a better outcome than outcome of diffuse midline H3K27M-mutant glioma. Our conclusion was not only that brainstem subependymomas can have H3K27M mutations, but that they do not carry the rapidly lethal prognosis with which these muta- tions are usually associated because of their discov- ery in diffuse intrinsic pontine gliomas.
Key words: H3K27M mutation; subependymoma; brain stem neoplasm; Sanger sequencing
Introduction
Subependymomas are rare primary central nervous system (CNS) tumors that occur mostly in middle-aged and elderly men. They account for 0.07%-0.51% of brain tumors, and are considered grade I benign tumors in the 2016 revision of the WHO classification of CNS tumors. Long-term survival is generally excellent even if the entire subependymoma cannot be removed which is said in reference 4 and 5. .The most common locations are in one of the lateral ventricles , but they also occur in the fourth ventricle, within the brain parenchyma, and in the spinal cord . Rare subependymomas are found in the brainstem. Subependymoma in brainstem exhib- ited a poorer prognosis. However, it remains unclear whether this poor prognosis is due to their location-related incomplete resection or due to their molecular features.
H3K27M glioblastomas are located in the midline and have been shown to have worse progno- sis than other GBM subgroups, with a median surviv- al of 6 months. Recent case reports have described the H3K27M mutation in certain midline low-gradetumors, such as pilocytic astrocytomas and other glioneuronal tumors, which suggests that H3K27M mutations may be present in a spectrum of low-grade glial tumors and mixed glioneuronal tumours, locat- ed in midline areas without necessarily signifying a poor prognosis. The frequency of H3K27M mutation in subependymoma, especially in the brain stem subependymoma, is unknown. In this study, we searched for H3K27M mutations in a series of 24 diagnosed subependymomas, using a combination of immunohistochemistry (IHC) and Sanger sequenc-ing.
Material and methods
Tumor samples
We studied 24 cases of grade I subependymo-ma, as defined by the WHO classification criteria between March 2008 and November 2017. We searched their files for cases of subependymoma and retrieved slides and blocks for 24 cases. All cases were independently reviewed by two pathologists of SanBo Brain Hospital. Disagreements in evaluation were resolved by review and discussion at a multi- headed microscope. The slides verify the diagnoses as subependymoma by the WHO 2016 criteria. All the selected cases presented typical features of subependymomas, which are the loose clusters of small bland round nuclei in a dense fibrillary matrix of glial cell processes with frequent occurrence of small cysts and occasionally exhibiting pseudoro- settes. Sections for IHC and genetic analyses were prepared from formalin-fixed paraffin embedded tissue specimens.
A range of other brain tumors was also investigat- ed and included the ependymomas (WHO Grade Ⅱ) (n = 18) and the anaplastic ependymomas (WHO Grade Ⅲ) (n = 13). The clinical characteristics of the patients with subependymomas are summarized in Table 1.
IHC analyses of H3F3A, HIST1H3B and HIST1H3C
Representative formalin-fixed sections were deparaffinized and stained with hematoxylin and eosin stained (HE) and IHC according to immuno-histochemical reagent instructions. In brief, 4μm sections were deparaffinized. The sections were then treated with 3% H2O2 for 5 min at room temperature to block endogenous peroxidase activity. For antigen retrieval, slides were pretreated by steaming in sodium citrate buffer for 15 min at 100°C. Then, the slides were blocked with 5% fetal bovine serum (FBS) at room temperature for 15 minutes and incu-bated with primary antibodies against GFAP(Dako,1:150), Olig-2 (Dako,1:100), EMA (Leica,1:100), Ki-67 (Dako,1:100), IDH1-R132H (Dianova, Germany,1:60), ATRX (Abcam, 1:800),TP53 (Santa Cruz Biotechnology, 1:200), H3K27M (EMD Millipore, 1:3000), and H3K27me3 (Cell Signaling Technology, 1:300) at 4°C over night. After being washed with PBS buffer, the sections were covered by anti-mouse/rabbit polymer HRP-label for 30min. Then DAB was applied for color development at room temperature for 5 minutes, and sections were subsequently counterstained with hematoxylin. Each slide was individually reviewed and scored by two experienced neuropathologists. In case of disagree-ment, the slide was re-examined and a consensus was reached by the observers. Ki67 was scored as the percentage of nuclei-stained cells out of all tumor cells in × 400 high-power field, about 1000 tumor cells were counted in overall for an entire slide.Adequate positive and negative controls were stained in parallel. Clinical features were obtained by reviewing the respective clinical records or by contacting the referring physicians. The study was approved by the Institutional Review Board of the SanBo Brain Hospital.
Mutation analyses of H3F3A, HIST1H3B and HIST1H3C
DNA extraction
Areas of the specimens that were enriched in tumor cells were marked on the H&E-stained sections. Tissue was scraped from this preselected area and transferred to an Eppendorf tube for DNA isolation using the QIAamp?DNA Mini kit (Qiagen GmbH, Germany) in accordance with the manufac-turer's protocol. The quality and concentration of DNA samples was examined using spectrophotome-ter (Biophtometer Eppendorf, Germany).
Sanger sequencing
Histone H3F3A, HIST1H3B and HIST1H3C were analyzed by direct sequencing of PCR-ampli- fied products from tumor DNA using the primers previously described. The amplified products were studied by direct sequencing after clean-up exonucle-ase ExoSAP-IT (Affymetrix, Santa Clara, CA) using the Big Dye Terminator Cycle Sequencing Kit and capillary electrophoresis on the automated sequencer ABI3730 (Applied Biosystems,Carlsbad, CA). Sense and antisense sequences were screened for exonic alterations using SeqScape v2.5software (Applied Biosystems) and compared with the National Center for Biotechnology Information (NCBI) reference sequences of H3F3A, HIST1H3B and HIST1H3C.
BRAFV600E Mutation Analysis
DNA was quantified to a final concentration of 2 ng/μl. BRAFV600E mutations were analyzed with an amplification-refractory mutation system using AmoyDx BRAF V600E Mutation Detection Kit (Amoy Diagnostics), according to the manu-facturer's instructions. The FAM fluorescence signal was used to evaluate the mutation status of the sample. When the sample FAM Ct value was ≥ 28, the sample was classified as negative or being below the detection limit of the kit. When the sam- ple FAM Ct value was <28, the sample was classi-fied as mutation-positive.
Fluorescence in situ hybridization (FISH)
Formalin-fixed, paraffin-embedded (FFPE) blocks were analyzed via FISH, using the techniques previously described. Briefly, 5-μm-thick FFPE sections were incubated at 56°C for 2 h, dewaxed, air dried, and dehydrated. Specimen were boiled in heat pretreatment solution for 40 min at 90°C, washed in 2X saline sodium citrate (SSC), digested with the enzyme reagent for 10 min at 37°C, washed in 2XSSC, dehydrated and air dried. The KIAA1549 (Agilent Technologies, USA, Green)/BRAF(Agilent Technologies, USA, red) probe mix was applied to the selected hybridization area for the fusion gene product of KIAA1549 and BRAF. Dual-color FISH was applied to the selected hybridization area using: locus specific identifier (LSI) 1p36/LSI 1q25 (Vysis/Abbott Molecular) and LSI 19q13/19p 13 dual-color probe (Vysis/Abbott Molecular) for loss of 1p36/19q13. The selected hybridization area was covered with a coverslip and sealed with rubber cement. DNA co-denaturation was performed at 85°C for 5 min and hybridization was allowed to occur at 38°C for 14-18h. Post-hybridization washes were performed by incubating in 2×SSC at 37°C for 10 min then 2×SSC/0.1% NP-40 at room temperature for 5min, followed by dehydration. Finally, 4',6-di- amidino-2-phenylindole (DAPI) was applied and the area covered with a coverslip.
Tumors were scored as positive for the BRAF-KIAA1549 fusion when >20% of the cells demonstrated yellow signals (indicating overlap of the green and red signals), in at least 100 non-over- lapping, intact nuclei. A total number of signals was counted and a ratio of 1p:1q (or 19q:19p) of <0.75 was diagnosed as loss (losses of 1p36/19q13).
Results
Clinical characteristics
Subependymoma
The clinical characteristics of the cases with subependymoma studied here are summarized in Table 1. The study cohort was comprised of 18 (75%)males and 6 (25%) females, with a median age at surgery of 45 years (range: 13-61 years). Twelve tumors were in a lateral ventricle, 5 were in the spinal cord, 4 in the brainstem, and 3 in temporal lobe.
Ependymomas (WHO grade Ⅱ and WHO grade Ⅲ)
The clinical characteristics of the 18 patients with grade II ependymoma and 13 patients with grade III anaplastic ependymoma are summarized in Table 1. The grade II ependymomas were from 15 males and 3 female, with a median age at surgery of 40.29 years (range: 12-61 years). Ten tumors were in the spinal cord, five were in the fourth ventricle, two in the frontal lobe, and one in the brainstem.
The grade III anaplastic ependymomas were from 8 males and 5 female, with a median age at surgery of 12.1 years (range: 2-31 years). Five of these were in the fourth ventricle, and 2 each were in the cerebellum, spinal cord, frontal lobe, and 1 each was in temporal lobe, and brainstem.
Pathological characteristics
The histological characteristics of the subep- endymoma in all 24 cases contained a coarse fibrillar matrix, with clusters of small uniform nuclei. Micro-cysts were common and were found in 18 (75%) tumors. None of the cases contained necrosis and microvascular proliferation. These tumors exhibited low-level mitotic activity, and the Ki-67 was less than 5% in any of tumors (Table 2). All of the tumors were immunopositive for GFAP. All had nuclear ATRX immunoreactivity (and so there was no ATRX muta-tion). No subependymoma was immunopositive for Olig2, TP53, or the protein product of the IDH1 R132H mutation. Eight had EMA immunoreactivity (Table 2).
H3K27M status was determined by IHC and Sanger sequencing. H3K27M immunoreactivity was only observed in the 4 subependymomas from the brainstem (Table 1, Table 2, and Figure. 1E-H). The histological evaluation showed typical morphology of subependymoma (Figure 1A-D). All 4 of the H3K27M immunopositive tumors were confirmed to have the mutations by Sanger sequencing (H3F3A K27M, AAG>ATG) (Figure 2A to 2D). No mutation in HIST1H3B or HIST1H3C gene was found. In the tumors with H3K27M mutations, there was loss of immunoreactivity for H3K27Me3 (Figure 1I-L). FISH for 1p/19q co-deletion and BRAF-KIAA1549 fusion and amplification-refractory mutation system methods for detecting BRAFV600E mutation were available for 4 brain cases. BRAFV600E mutation and BRAF-KIAA1549 fusion (Figure 3I-L) were not found in 4 patients. No 1p/19q co-deletion was detect- ed in 4 cases (Figure 3A-H).
H3K27M mutations were not found in any of the grade II or grade III ependymomas (Table 1). Tumors without H3K27M mutations all displayed H3K27me3.
Clinical features of the 4 H3K27M-mutant brain-stem subependymomas
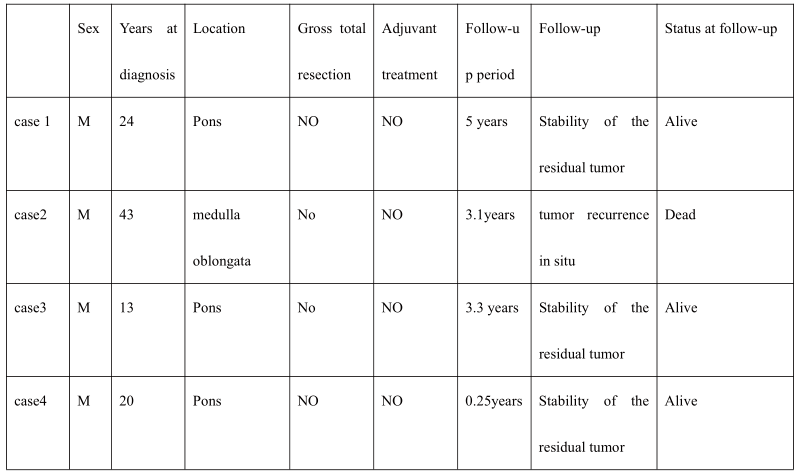
The clinical and radiological characteristics of the 4 patients with H3K27M mutations are summa- rized in Table 3 and Figurer 3. These patients had a median age of 22 years (range: 13-43 years). All 4 patients were male. The tumors were located in the medulla oblongata and pons. The tumors in case 1, 3, and 4 were removed through right frontotempo- ral-subtemporal approach. The tumor in case 2 was removed through posterior median approach. No gross total resection was performed. The patients are still alive at 60, 39.6, and 3 months post-surgery, and one died at 37.2 months. The median OS, defined by the follow-up period, was 3.2 years (range: 0.25-5 years). None of the patients received postsurgical treatment (radiotherapy and chemotherapy).
These tumors were all isointense to gray matter in T1-weighted MR images (Figure 4A, 4E and 4I) and were slightly hyperintense compared to gray matter in T2-weighted (Figure. 4B, 4F and 4G) and FLAIR sequences (Figure 4C and 4H), but contrast-enhanced MRI showed no enhancement (Figure 4D, 4G, 4H and 4I).
Discussion
We have described 4 cases of brainstem subep- endymoma with H3K27M mutations, whereas 20 others from other CNS sites lacked such mutations. Ours is the first report of such mutations in subep- endymomas. All 4 of the tumors were histologically ordinary grade I examples without any features of high-grade gliomas, and 3 of the 4 patients are still alive without evidence of progressive disease despite having undergone only partial resections without any adjunctive therapy. This finding is in considerable contrast to the survival of diffuse midline gliomas with H3K27M mutations, and is similar to the report- ed good survival of a child with a pilocytic astrocyto- ma with such a mutation or the long survival report- ed for some patients with midline gangliogliomas or other glioneuronal tumors with such mutations.
The H3K27M mutation has been described in midline low-grade glial tumors and glial-neuronal tumors , even in 2 cases of posterior fossa ependymoma . All together, these data suggest that H3K27M mutation is not specific to malignant grade IV glial tumor. In agreement with the evaluated H3K27M expression, we identified that a global reduction in H3K27me3 in tumors with H3K27M mutation compared to tumors without H3K27M mutation, which is in line with previous reports . Four cases of subependymoma with H3K27M mutation did not possess glioma’s molecular pathologic characteristics, for instance, KIAA1549-BRAF fusion, BRAF V600E mutation, IDH1R132H, TP53 mutation, ATRX mutation and 1p19q co-delection. And no H3K27M mutations were found in any case of brainstem ependymoma whether in this cohort or in literature. Our study suggests that nuclear H3K27M expression may potentially be a sensitive and specific marker for brainstem subependymoma, which may be helpful in differentiating patients with ependymoma in brainstem.
It must be noted that our study is limited to a relatively small number of subependymomas and ependymo- mas (grade Ⅱ and grade Ⅲ). For this reason, our results are suggestive but not conclusive for clinical practice, with respect to the higher sensitivity of H3K27M mutation in the differential diagnosis of brainstem subependy- moma vs. brainstem ependymoma.
Conflict of Interest:
Yao Kun, Duan Zejun, Yin Wang, Mingshan Zhang, Tao Fan, Bin Wu and Qi Xueling each declares that they have no conflict of interest.
Ethical approval:
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional.
This article does not contain any studies with animals performed by any of the authors.
Informed consent:
Informed consent was obtained from all individual participants included in the study.This article does not contain any studies with animals performed by any of the authors.
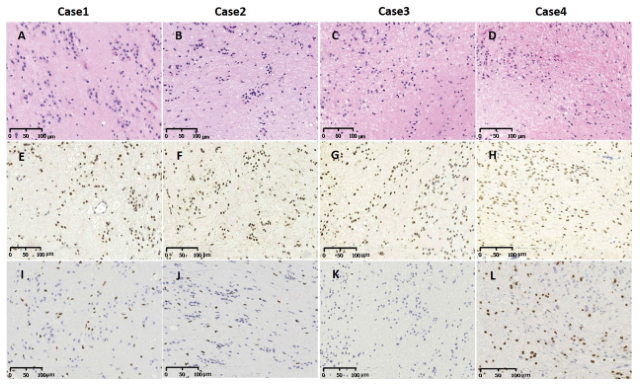
Figure 1. Histological and immunohist ochemical analyses of 4 cases of subependymoma in brain stem with the H3K27M mutations. H&E revealed the diagnosis of subependymomas (Figure A-D).Tumors cells showed magnification 100. IHC staining of 4 cases of subependymomas in brain stem using anti-H3K27M (Figure E-H) and anti-H3K27me3 (Figure I-L) antibodies. H3K27M shows strong nuclear positivity in tumor cell, but no staining in the nuclei of endothelial and smooth muscle cell in blood vessels (Figure E-H). Accordingly, H3K27me3 staining on the same samples shows loss of the expression of this histone marker in H3K27M mutant tumors (Figure I-L).
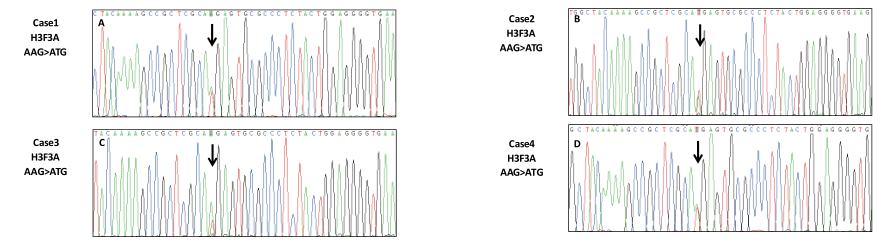
Figure 2. Mutation analysis of H3F3A in cases of 1-4 showing G> T mutation (Figure A-D)Sanger sequencing chromatograph of resulting PCR-amplified H3F3A confirmed c.83A > T transversion in DNA of cases of 1-4 tumor tissue (arrow).
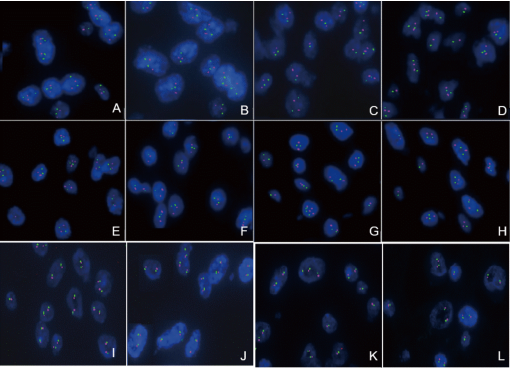
Figure 3. FISH performed on the 4 cases for detecting the chromosome 1p36 and 19q13 and for the BRAF-KIAA1549 fusion.Case1 tumor cells showed no deletion of 1p36 (Figure A) and 19q13 (Figure B); Case2 tumor cells showed no deletion of 1p36 (Figure C) and 19q13 (Figure D); Case3 tumor cells showed no deletion of 1p36 (Figure E) and 19q13 (Figure F); Case4 tumor cells showed no deletion of 1p36 (Figure G) and 19q13 (Figure H). Case1 (Figure I), case2 (Figure J), case3 (Figure K) and case4 (Figure L) tumor cells showed no BRAF-KIAA1549 fusion.
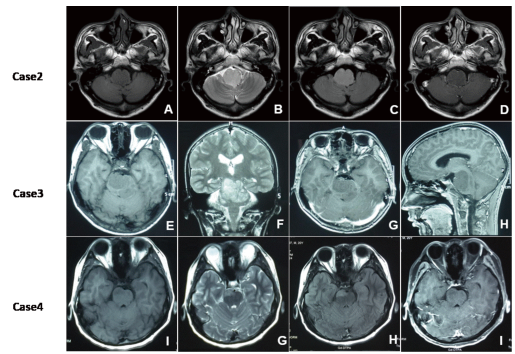
Figure 4.The preoperative radiologic imaging of case 2,3,4;These tumors were all isointense to gray matter in T1-weighted MR images (Figure A, E and I) and were slightly hyperintense compared to gray matter in T2-weighted (Figure. B, F and G) and FLAIR sequences (Figure C and H), but contrast-enhanced MRI showed no enhancement (Figure D, G, H and I).
Table 1. Clinical characteristics of the subependymomas and grade II or grade III anaplastic ependymomas examined in this study
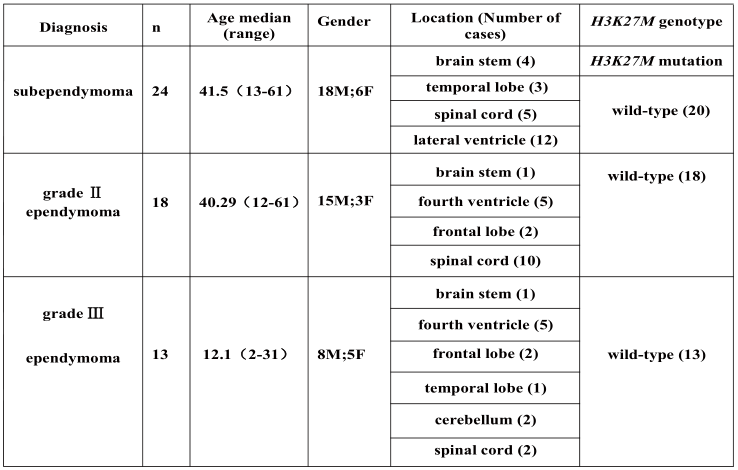
Table 2. Histological characteristics of the 24 subependymomas examined in this study
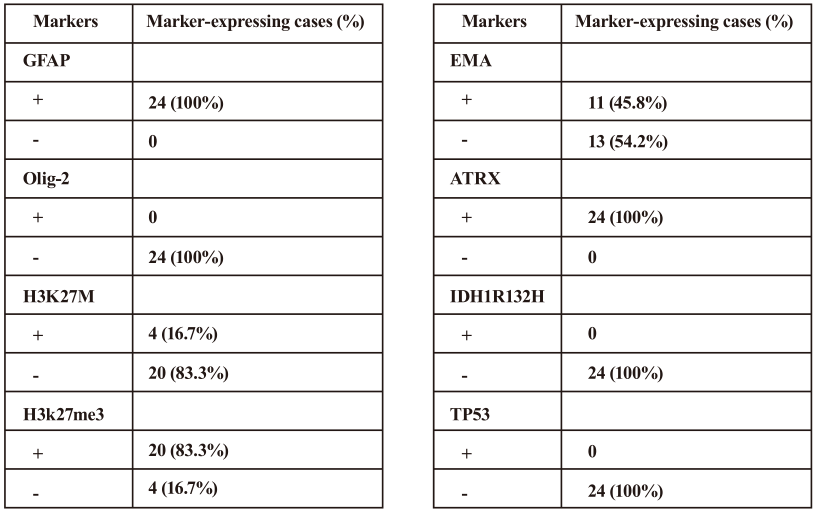
The expression levels were based on the percentages of the immunopositive cells(negative, < 10% of tumor cells; positive, ≥10% of tumor cells).
Table 3. Clinical features of the 4 cases of subependymoma with H3K27M mutations




 京公網(wǎng)安備 11010802035500號(hào)
京公網(wǎng)安備 11010802035500號(hào)